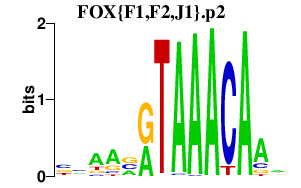
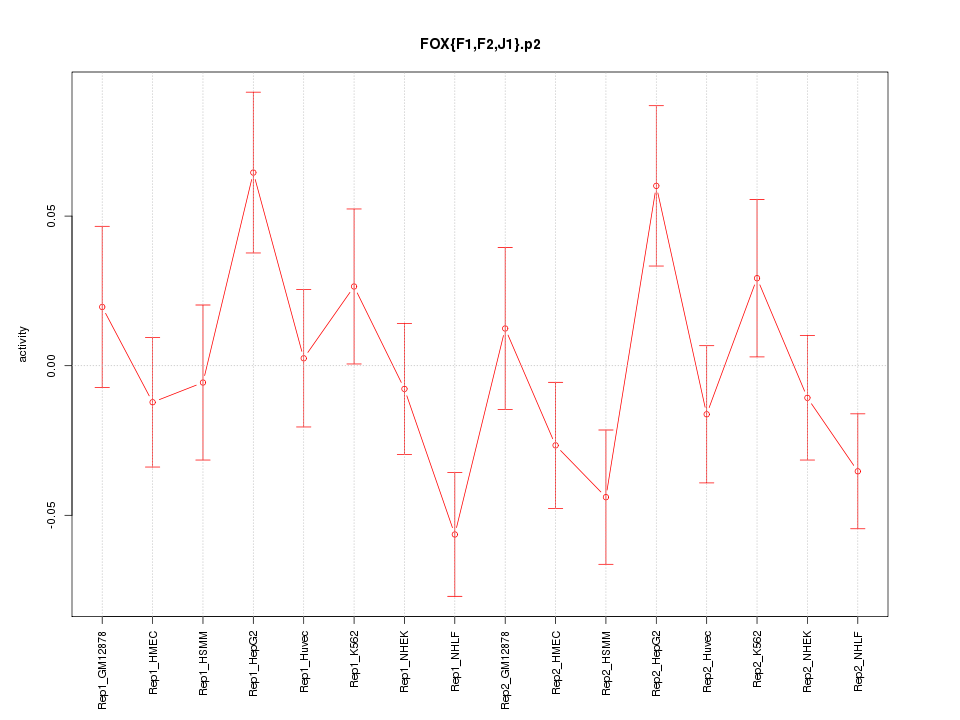
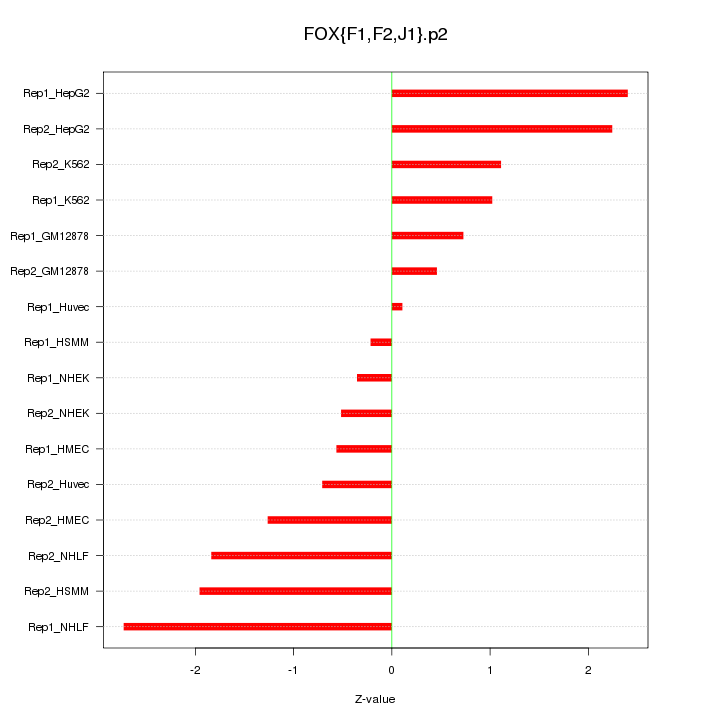
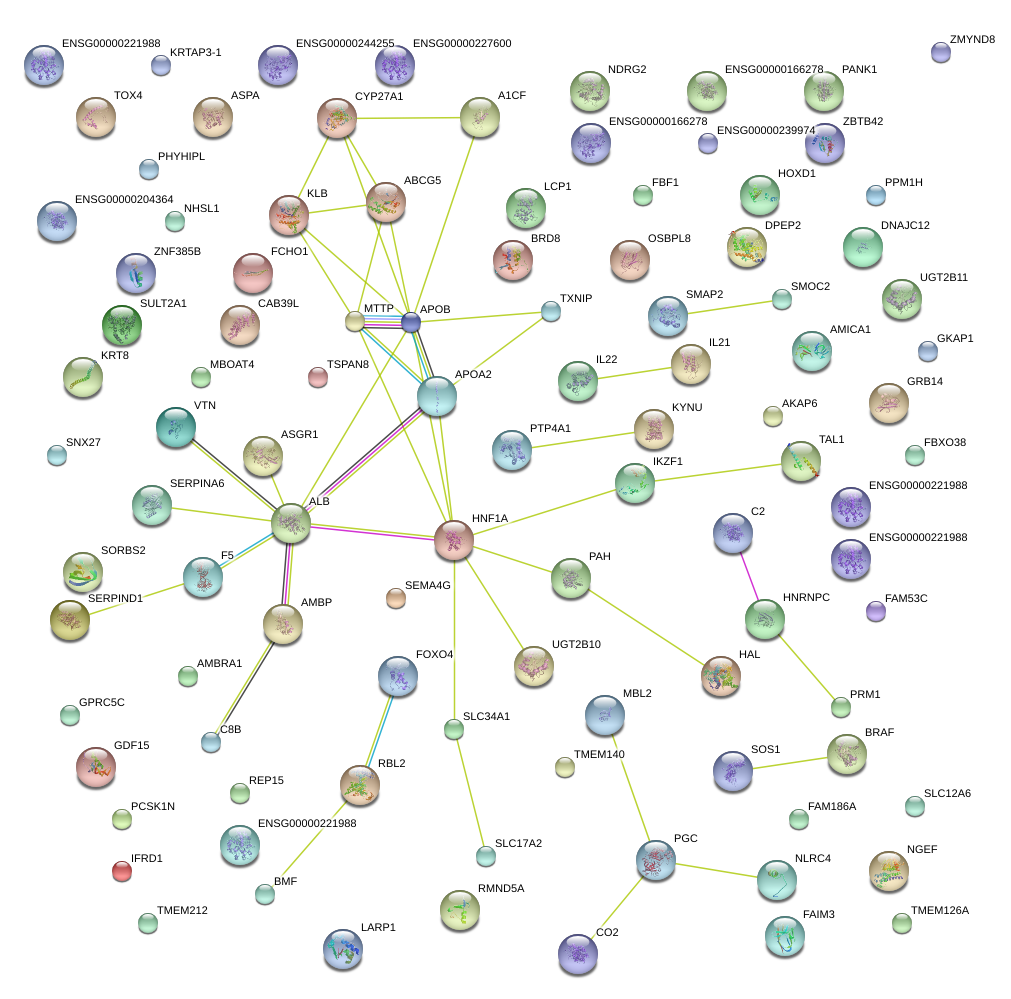
|
chr1_-_159459814
|
4.056
|
|
APOA2
|
apolipoprotein A-II
|
|
chr1_-_159459999
|
4.042
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr11_-_117589283
|
3.030
|
NM_153206
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1
|
|
chr7_+_134483305
|
3.026
|
NM_018295
|
TMEM140
|
transmembrane protein 140
|
|
chr5_-_42847716
|
2.928
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr22_+_19458378
|
2.861
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr17_+_69939130
|
2.817
|
NM_022036
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr14_-_20562962
|
2.612
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr17_-_7023283
|
2.580
|
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr12_-_51629874
|
2.417
|
|
KRT8
|
keratin 8
|
|
chr20_-_45410033
|
2.414
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr16_-_11282595
|
2.351
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr6_-_41823056
|
2.224
|
NM_001166424
NM_002630
|
PGC
|
progastricsin (pepsinogen C)
|
|
chr3_+_173043832
|
2.147
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr1_-_57204208
|
2.087
|
NM_000066
|
C8B
|
complement component 8, beta polypeptide
|
|
chr13_+_49658016
|
2.073
|
|
|
|
|
chr2_-_233586145
|
2.059
|
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr17_-_7023378
|
2.054
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr2_-_233586164
|
2.023
|
NM_019850
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr2_-_165133239
|
2.003
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr10_-_91395194
|
1.967
|
NM_148977
|
PANK1
|
pantothenate kinase 1
|
|
chr12_+_119900677
|
1.883
|
|
HNF1A
|
HNF1 homeobox A
|
|
chr12_+_119900931
|
1.843
|
NM_000545
|
HNF1A
|
HNF1 homeobox A
|
|
chr4_+_100714995
|
1.828
|
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr1_+_116455898
|
1.821
|
NM_152367
|
MAB21L3
|
mab-21-like 3 (C. elegans)
|
|
chr6_-_26038817
|
1.712
|
NM_005835
|
SLC17A2
|
solute carrier family 17 (sodium phosphate), member 2
|
|
chr1_-_167822248
|
1.702
|
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chr17_-_36418869
|
1.659
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chr17_+_3324096
|
1.640
|
NM_001128085
|
ASPA
|
aspartoacylase
|
|
chr12_-_101835026
|
1.604
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr6_-_138862271
|
1.595
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr6_-_138862273
|
1.581
|
|
|
|
|
chr6_+_64289585
|
1.563
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr4_+_69716298
|
1.520
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr16_-_66590856
|
1.499
|
NM_022355
|
DPEP2
|
dipeptidase 2
|
|
chr19_+_18357966
|
1.477
|
NM_004864
|
GDF15
|
growth differentiation factor 15
|
|
chr1_-_167822334
|
1.466
|
NM_000130
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chr1_-_47468029
|
1.417
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr13_-_45654327
|
1.407
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr13_-_45654292
|
1.405
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr1_+_40635058
|
1.378
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr13_-_45654296
|
1.378
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr1_+_144149794
|
1.361
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr8_-_30121741
|
1.348
|
NM_001100916
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4
|
|
chr14_+_104338423
|
1.345
|
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr6_+_32003180
|
1.343
|
NM_000063
NM_001145903
|
C2
|
complement component 2
|
|
chr10_-_52315390
|
1.328
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr7_+_50318801
|
1.324
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr14_+_104337923
|
1.309
|
NM_001137601
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr5_+_147754467
|
1.298
|
|
FBXO38
|
F-box protein 38
|
|
chr14_-_93859394
|
1.269
|
NM_001756
|
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6
|
|
chr14_+_31868229
|
1.265
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr2_+_143351695
|
1.204
|
|
KYNU
|
kynureninase
|
|
chr12_-_69837831
|
1.197
|
|
TSPAN8
|
tetraspanin 8
|
|
chr2_-_39201631
|
1.191
|
|
SOS1
|
son of sevenless homolog 1 (Drosophila)
|
|
chr2_+_219354692
|
1.180
|
NM_000784
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1
|
|
chr19_-_53081326
|
1.164
|
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr12_-_61615124
|
1.143
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr10_+_102722275
|
1.141
|
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr19_+_17723285
|
1.135
|
NM_001161359
NM_015122
|
FCHO1
|
FCH domain only 1
|
|
chr15_-_32398220
|
1.133
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr12_-_94914133
|
1.125
|
NM_002108
|
HAL
|
histidine ammonia-lyase
|
|
chr12_-_49076658
|
1.115
|
NM_001145475
|
FAM186A
|
family with sequence similarity 186, member A
|
|
chr17_-_23721485
|
1.114
|
NM_000638
|
VTN
|
vitronectin
|
|
chrX_+_70232362
|
1.109
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr10_-_69267774
|
1.101
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr4_-_123761615
|
1.087
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr10_-_52315436
|
1.072
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr2_-_32344192
|
1.058
|
NM_021209
|
NLRC4
|
NLR family, CARD domain containing 4
|
|
chr11_-_46572073
|
1.055
|
|
AMBRA1
|
autophagy/beclin-1 regulator 1
|
|
chr4_-_186969041
|
1.039
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr2_-_43919392
|
1.030
|
NM_022436
|
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5
|
|
chr1_-_1346465
|
1.013
|
NM_001145210
|
LOC441869
|
hypothetical protein LOC441869
|
|
chr15_-_38185578
|
1.009
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr17_-_23721468
|
1.008
|
|
VTN
|
vitronectin
|
|
chr12_+_27740694
|
1.007
|
NM_001029874
|
REP15
|
RAB15 effector protein
|
|
chr1_-_205161821
|
0.996
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr5_+_154072654
|
0.971
|
NM_015315
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr17_-_23721430
|
0.968
|
|
VTN
|
vitronectin
|
|
chr11_-_46572160
|
0.952
|
|
AMBRA1
|
autophagy/beclin-1 regulator 1
|
|
chr10_-_54201400
|
0.950
|
NM_000242
|
MBL2
|
mannose-binding lectin (protein C) 2, soluble
|
|
chr10_+_60606352
|
0.949
|
NM_032439
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like
|
|
chr4_+_74488835
|
0.942
|
NM_000477
|
ALB
|
albumin
|
|
chr13_-_48873735
|
0.931
|
NM_030925
|
CAB39L
|
calcium binding protein 39-like
|
|
chr12_-_69838045
|
0.930
|
NM_004616
|
TSPAN8
|
tetraspanin 8
|
|
chrX_-_48578878
|
0.928
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr7_+_111850367
|
0.927
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr2_+_143351536
|
0.927
|
NM_001032998
NM_003937
|
KYNU
|
kynureninase
|
|
chr4_+_74488910
|
0.924
|
|
ALB
|
albumin
|
|
chr6_+_32230172
|
0.916
|
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr5_+_137701122
|
0.896
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr7_+_149733309
|
0.888
|
|
|
|
|
chr17_-_23721420
|
0.884
|
|
VTN
|
vitronectin
|
|
chr9_-_115880494
|
0.878
|
NM_001633
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr2_-_21084077
|
0.873
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr4_+_39084867
|
0.869
|
NM_175737
|
KLB
|
klotho beta
|
|
chr2_+_176761552
|
0.865
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr9_-_85622366
|
0.854
|
NM_001135953
NM_025211
|
GKAP1
|
G kinase anchoring protein 1
|
|
chr16_+_52027020
|
0.852
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr7_-_140270749
|
0.848
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr2_+_86800940
|
0.847
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr1_+_149877955
|
0.846
|
|
SNX27
|
sorting nexin family member 27
|
|
chr2_-_180319013
|
0.843
|
NM_001113397
|
ZNF385B
|
zinc finger protein 385B
|
|
chrX_+_46825179
|
0.842
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr14_+_94148466
|
0.841
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr3_+_99934227
|
0.831
|
NM_006100
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6
|
|
chr3_-_58497913
|
0.826
|
NM_003500
|
ACOX2
|
acyl-CoA oxidase 2, branched chain
|
|
chr2_+_86800898
|
0.819
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr17_-_23721351
|
0.818
|
|
VTN
|
vitronectin
|
|
chr9_-_115880366
|
0.813
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr6_+_32229599
|
0.809
|
NM_005155
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr20_-_22514092
|
0.805
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr4_-_186969202
|
0.805
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr7_+_106292975
|
0.805
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr7_+_1094248
|
0.800
|
NM_001098201
|
GPER
|
G protein-coupled estrogen receptor 1
|
|
chr18_+_53863865
|
0.791
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr1_+_190811479
|
0.785
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr16_+_11346780
|
0.782
|
NM_152308
|
C16orf75
|
chromosome 16 open reading frame 75
|
|
chr15_-_38185865
|
0.780
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr4_+_39084936
|
0.773
|
|
KLB
|
klotho beta
|
|
chr2_+_169637255
|
0.772
|
NM_001142270
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr4_-_155731288
|
0.768
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr19_-_47638890
|
0.767
|
NM_198477
|
CXCL17
|
chemokine (C-X-C motif) ligand 17
|
|
chr6_-_31740466
|
0.763
|
|
GPANK1
|
G patch domain and ankyrin repeats 1
|
|
chr9_-_115880281
|
0.761
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr5_-_59819642
|
0.760
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chrX_-_153698930
|
0.755
|
|
HMGN1P37
|
high-mobility group nucleosome binding domain 1 pseudogene 37
|
|
chr3_-_10522267
|
0.752
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr17_+_69950782
|
0.743
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr18_+_59726183
|
0.739
|
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr9_+_135276940
|
0.734
|
NM_139025
NM_139026
NM_139027
|
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13
|
|
chr5_+_137701601
|
0.731
|
NM_016605
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr15_-_38387357
|
0.728
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr1_+_111483771
|
0.720
|
NM_001007794
|
CEPT1
|
choline/ethanolamine phosphotransferase 1
|
|
chr15_-_38387410
|
0.717
|
NM_004573
|
PLCB2
|
phospholipase C, beta 2
|
|
chr8_+_82807218
|
0.715
|
NM_152284
|
CHMP4C
|
chromatin modifying protein 4C
|
|
chr1_-_24067347
|
0.696
|
NM_000147
|
FUCA1
|
fucosidase, alpha-L- 1, tissue
|
|
chr19_+_15612706
|
0.690
|
NM_000896
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3
|
|
chr9_+_132990760
|
0.690
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr11_+_22644732
|
0.687
|
NM_001143830
|
GAS2
|
growth arrest-specific 2
|
|
chr10_-_50066386
|
0.685
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr4_-_111339201
|
0.684
|
NM_001130721
NM_024090
|
ELOVL6
|
ELOVL family member 6, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr2_+_11591692
|
0.676
|
NM_014668
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr6_+_30238961
|
0.672
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr3_-_71376919
|
0.671
|
|
FOXP1
|
forkhead box P1
|
|
chr11_-_116213511
|
0.665
|
NM_000039
|
APOA1
|
apolipoprotein A-I
|
|
chr4_+_22608252
|
0.660
|
|
|
|
|
chr15_-_38387164
|
0.658
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr12_-_31370305
|
0.657
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr6_+_43135606
|
0.648
|
NM_201523
|
KLC4
|
kinesin light chain 4
|
|
chr17_-_73636386
|
0.640
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr2_+_113601608
|
0.636
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr5_+_137701852
|
0.636
|
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr5_-_169657360
|
0.634
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr17_+_70244945
|
0.633
|
NM_001006638
NM_001163990
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr19_-_51979838
|
0.629
|
NM_001145145
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5
|
|
chr2_+_43919606
|
0.628
|
NM_022437
|
ABCG8
|
ATP-binding cassette, sub-family G (WHITE), member 8
|
|
chr2_-_39310176
|
0.625
|
NM_001009565
|
CDKL4
|
cyclin-dependent kinase-like 4
|
|
chr2_+_201756048
|
0.623
|
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr5_-_169657395
|
0.621
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr8_-_110724594
|
0.620
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr12_-_31370351
|
0.617
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr14_+_20226762
|
0.617
|
NM_001097577
NM_194431
|
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5
ribonuclease, RNase A family, 4
|
|
chr9_-_115880372
|
0.614
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr6_+_17708483
|
0.610
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr5_-_169657312
|
0.609
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr4_+_187424111
|
0.609
|
NM_000128
|
F11
|
coagulation factor XI
|
|
chr6_+_154373323
|
0.604
|
NM_001145279
NM_001145280
NM_001145281
|
OPRM1
|
opioid receptor, mu 1
|
|
chr12_-_31370387
|
0.603
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr16_+_11346812
|
0.600
|
|
C16orf75
|
chromosome 16 open reading frame 75
|
|
chr6_-_43135092
|
0.599
|
NM_015950
|
MRPL2
|
mitochondrial ribosomal protein L2
|
|
chr10_+_7785347
|
0.598
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr1_-_229627412
|
0.594
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr10_+_5126589
|
0.591
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr5_+_137701855
|
0.591
|
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr11_-_102331643
|
0.590
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr15_+_21361546
|
0.585
|
NM_005664
|
MKRN3
|
makorin ring finger protein 3
|
|
chr2_-_128332143
|
0.579
|
|
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D
|
|
chr18_-_50777634
|
0.578
|
NM_025214
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr17_+_75366524
|
0.577
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr17_-_6958695
|
0.573
|
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr2_+_113601616
|
0.572
|
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr2_+_168383427
|
0.567
|
NM_020981
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr6_+_32229771
|
0.567
|
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr10_+_114144512
|
0.566
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr2_+_86801081
|
0.565
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr4_+_74491272
|
0.558
|
|
ALB
|
albumin
|
|
chr11_-_114880275
|
0.555
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr12_+_120726317
|
0.549
|
|
SETD1B
|
SET domain containing 1B
|
|
chr4_+_187424336
|
0.544
|
|
F11
|
coagulation factor XI
|
|
chr10_+_7785331
|
0.544
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr1_+_196874867
|
0.543
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr12_+_9033403
|
0.539
|
NM_005810
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1
|
|
chr3_-_10522340
|
0.539
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr7_+_106293112
|
0.536
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr4_+_71096830
|
0.535
|
NM_017855
|
ODAM
|
odontogenic, ameloblast asssociated
|
|
chr12_+_52121699
|
0.532
|
NM_001005354
NM_018457
|
PRR13
|
proline rich 13
|
|
chr2_-_128332161
|
0.531
|
|
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D
|
|
chr17_-_72151314
|
0.526
|
NM_018414
|
ST6GALNAC1
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1
|
|
chr2_-_87156616
|
0.525
|
|
LOC285074
|
anaphase promoting complex subunit 1 pseudogene
|